SciToolAgent: Integration of 500+ research tools and automation of research and scientific tasks for intelligent bodies
General Introduction
SciToolAgent is an open source tool platform developed by the Innovation Center of Zhejiang University in Hangzhou (HICAI-ZJU). It integrates more than 500 scientific tools through knowledge graph (SciToolKG) and big language modeling technologies to help researchers deal with problems in the fields of biology, chemistry, and materials science. The main use of this tool is to automate scientific research tasks, such as data processing, molecular modeling or literature analysis, and ultimately to generate specific results, such as analytical charts, computational results or summary reports. The platform is hosted for free on GitHub and can be deployed locally or experienced through an online service, making it particularly suitable for research teams that need to produce results quickly.

Function List
- Tool Integration: Connect more than 500 tools to handle data, modeling or computational tasks.
- Knowledge Graph Recommendations: Recommend the right combination of tools based on the needs of the task.
- Task automation: Save time by automatically planning and executing research processes.
- Results generation: Output specific results such as graphs, reports or calculations.
- safety monitoring: Ensure that the tool runs safely and avoids erroneous results.
- Custom extensions: Support the addition of new tools to produce personalized results.
Using Help
Installation process
SciToolAgent requires a local installation to be fully functional. The steps are as follows:
1. Downloading the source code
- Enter it in the terminal:
git clone https://github.com/HICAI-ZJU/SciToolAgent.git
cd SciToolAgent
2. Creation of virtual environments
- Create and activate the environment:
conda create -n SciToolAgent python=3.10
conda activate SciToolAgent
3. Installation of dependencies
- Install the full dependency:
pip install -r requirements.txt
- Or install the lite version:
pip install -r requirement_agent.txt
4. Configuration environment
- Copy and edit
.envDocumentation:
OPENAI_API_BASE = your_api_base
OPENAI_API_KEY = your_api_key
5. Activation of services
- go into
toolsCatalog, run:
cd tools
bash run.sh
How to use
SciToolAgent can output results either by running locally or experiencing them online.
local operation
- Starting services: Run
bash run.shAfter that, the service is on standby locally. - operate: Enter
testdirectory, run the test script:
cd ../test
PYTHONPATH=. python test_run_SciToolAgent.py
- View Results: After the task is completed, the results are saved in the
tools/TempFiles, such as charts or reports. - running case: Root directory
Cases.ipynbFour examples are provided and can be run directly to view the output.
Online Experience
- Access to online services:
http://scitoolagent.scimind.ai:8080/ - Enter a task (e.g. "Analyze molecular data") and return simple results within seconds of submission.
- The online version is suitable for quick testing, complex results need to be run locally.
Featured Functions Operation Procedure
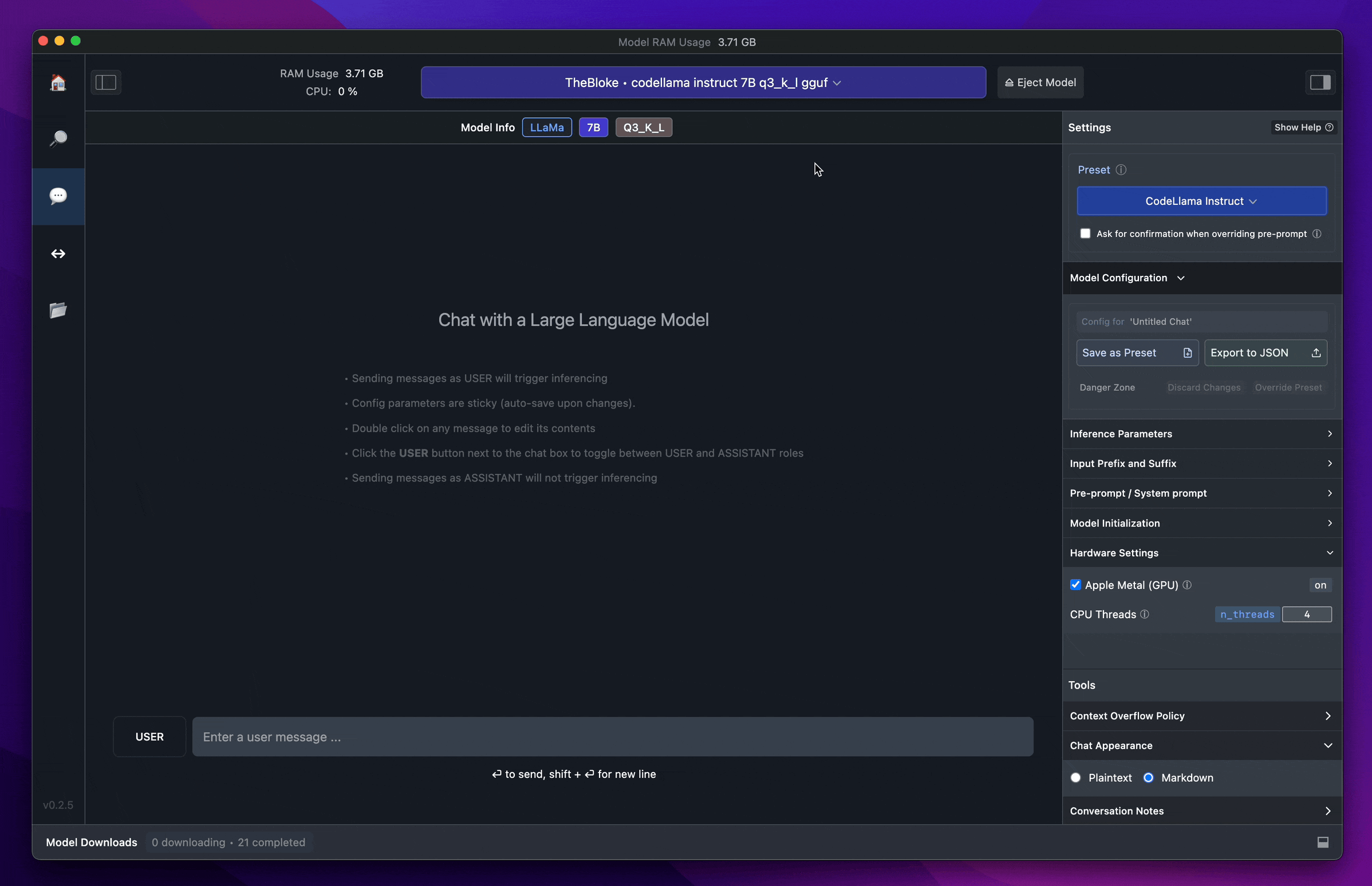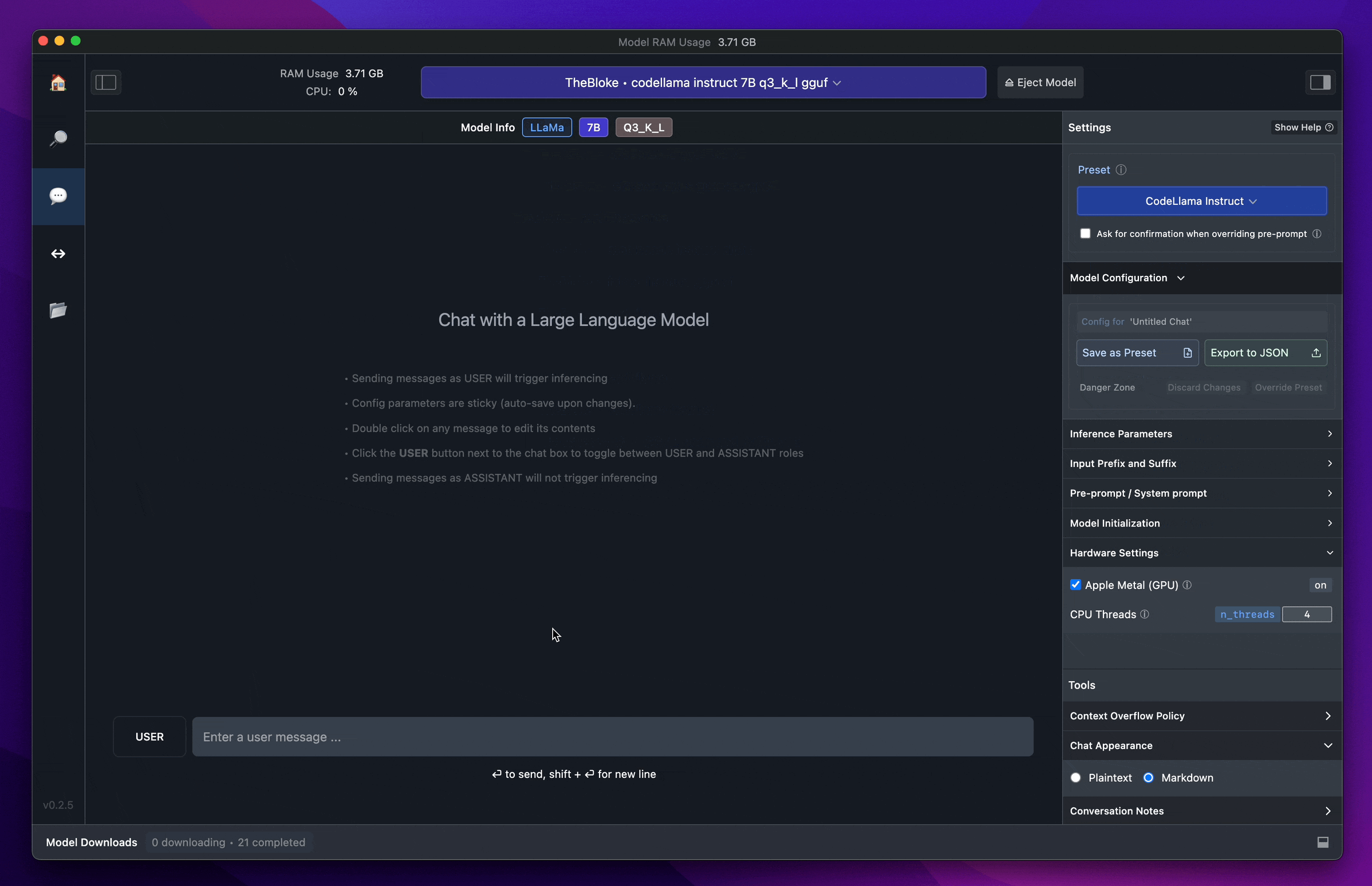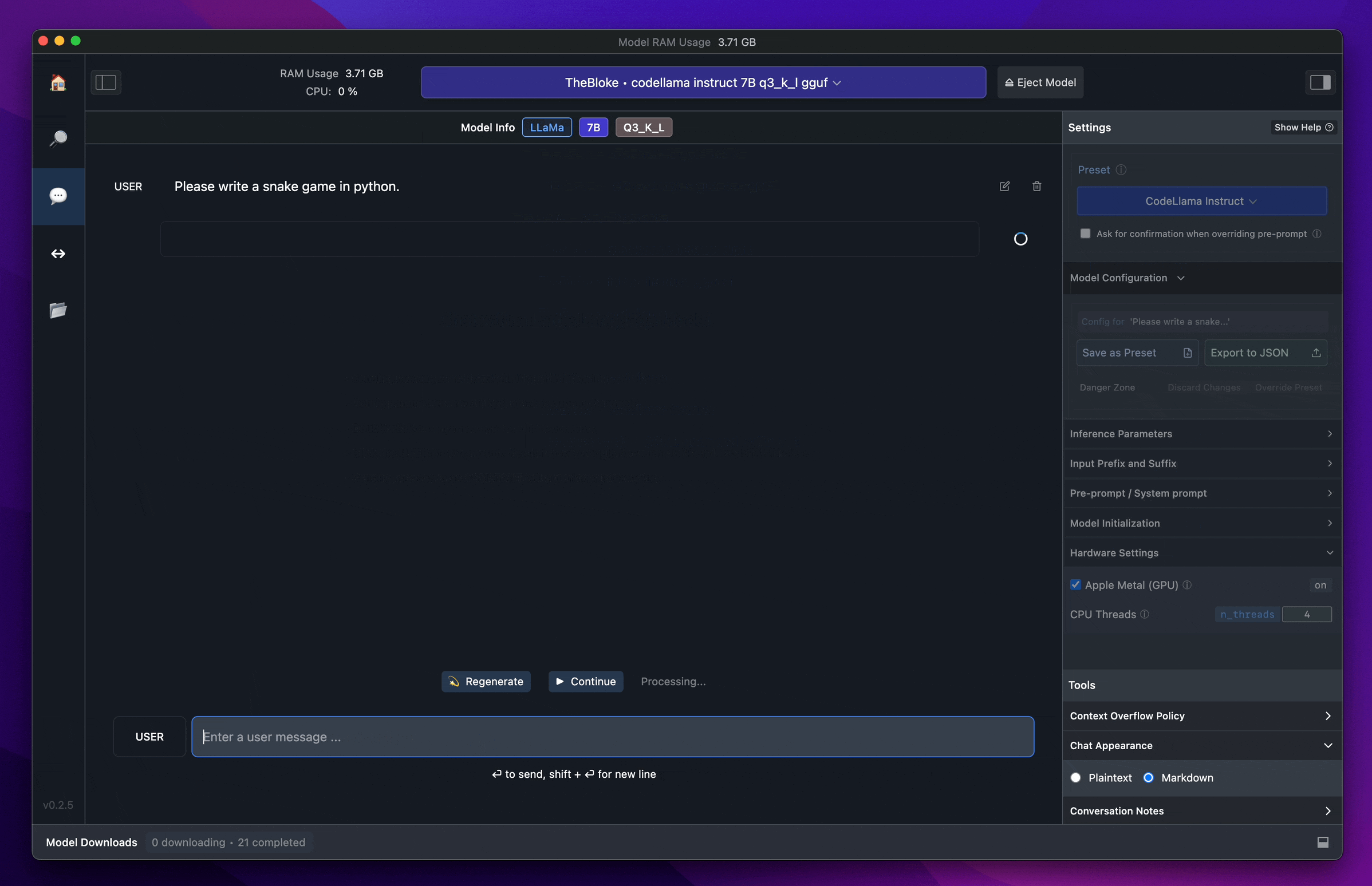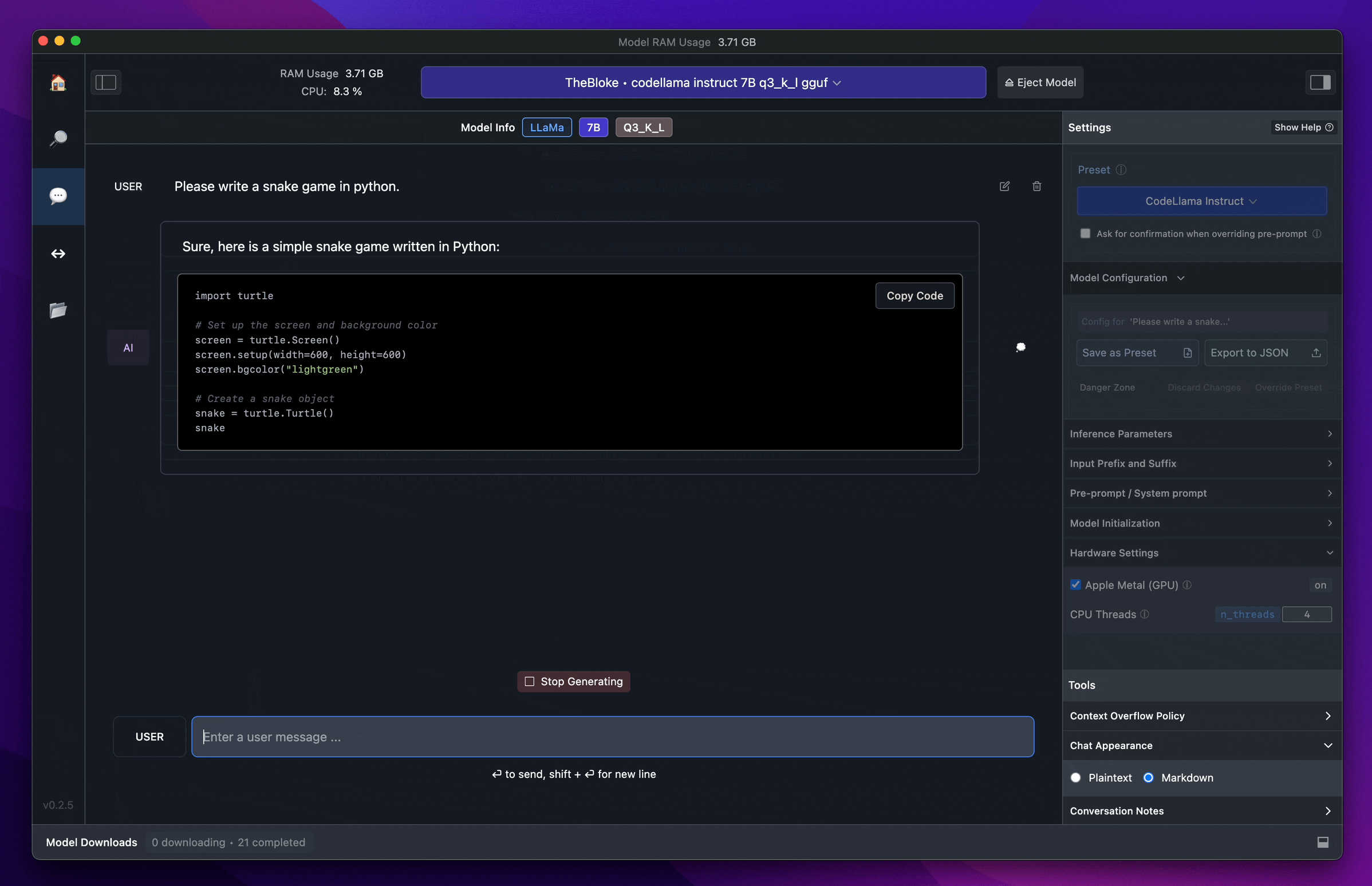
1. Data analysis and graphical output
- use: Processing data and generating visualizations.
- move::
- To convert a data file (e.g.
.csv(computing) put (sth) into (the)tools/DataFilesThe - Run command:
python scripts/run_agent.py --input "tools/DataFiles/sample.csv" --task "数据分析并可视化"
- probe
tools/TempFilesThe following is a list of the charts and analysis results that are generated.
2. Molecular modeling to generate calculations
- use: Analyze molecular structures and output calculated data.
- move::
- Prepare molecular documents (e.g.
.cif), put in thetools/DataFilesThe - Calling Tools:
from tools.ToolsFuns import molecular_analysis
result = molecular_analysis("tools/DataFiles/input_file.cif")
print(result)
- The output results are molecular parameters or model data, saved in the specified path.
3. Automated generation of scientific reports
- use: Integrate multiple tools to output complete reports.
- move::
- Enter the task description and data file:
python scripts/run_agent.py --input "tools/DataFiles/sample.pdf" --task "分析文献并生成报告" - The system automatically invokes the tool, processes the literature and generates a summary.
- The report is kept in the
tools/TempFilesThe format is text or PDF.
caveat
- Local operation requires 16GB of RAM and a stable network to ensure successful completion of complex tasks.
- The online version is suitable for simple results, and local deployment is recommended for large amounts of data.
- Regular cleaning
tools/TempFilesto avoid taking up too much space.
© Copyright notes
Article copyright AI Sharing Circle All, please do not reproduce without permission.
Related posts

No comments...