AI-Scientist-v2: Autonomous completion of scientific research and paper writing
General Introduction
AI-Scientist-v2 is an intelligent system developed by the Japanese company SakanaAI that aims to automate scientific research from start to finish by machines. It can come up with research ideas, design experiments, run code, analyze data, and finally write scientific papers. in April 2025, the tool was open sourced on GitHub, upgraded with a first version that added Agentic Tree Search technology to make exploration smarter. The first paper it generated that was written entirely by AI has passed peer review at the ICLR 2025 workshop.AI-Scientist-v2 does not rely on human templates and is applicable to a wide range of machine learning domains, making it suitable for researchers and developers.
Function List
- Presentation of research ideas: Automatically generate feasible research ideas based on input directions.
- Write experimental code: Generate the code needed to run experiments, support tuning and optimization.
- Execution of experiments and analysis: Automatically runs code, collects data, and generates charts.
- Writing scientific papers: Output a well-formatted paper based on the results of the experiment.
- Intelligent Path Optimization: Explore the best research options through Agentic Tree Search.
- Literature Search Support: Optional access to the Semantic Scholar API to check for novelty and add citations.
- open source: Full code is provided and users are free to modify and extend it.
Using Help
AI-Scientist-v2 requires a certain amount of technical knowledge, but when configured it can dramatically simplify scientific research. Below are detailed steps to help users get started quickly.
Installation process
- Preparing the environment
- Requires Linux and an NVIDIA GPU with CUDA and PyTorch support.
- Create a Python 3.11 environment:
conda create -n ai_scientist python=3.11 conda activate ai_scientist - Install PyTorch and CUDA:
conda install pytorch torchvision torchaudio pytorch-cuda=12.4 -c pytorch -c nvidia
- Installation of dependencies
- Download code:
git clone https://github.com/SakanaAI/AI-Scientist-v2.git cd AI-Scientist-v2 - Install additional tools:
conda install anaconda::poppler # 处理 PDF conda install conda-forge::chktex # 检查论文格式 pip install -r requirements.txt
- Download code:
- Configuring the API
- Set the big model API key (e.g. OpenAI):
export OPENAI_API_KEY='你的密钥' - If you use Claude models, installing additional support:
pip install anthropic[bedrock]Set up AWS keys and regions:
export AWS_ACCESS_KEY_ID='你的ID' export AWS_SECRET_ACCESS_KEY='你的密钥' export AWS_REGION_NAME='us-west-2' - Optionally configure the Semantic Scholar API:
export S2_API_KEY='你的密钥'
- Set the big model API key (e.g. OpenAI):
- test environment
- Check if the GPU is available:
python -c "import torch; print(torch.cuda.is_available())" - exports
TrueIndicates successful installation.
- Check if the GPU is available:
How to use the main features
1. Generating research ideas
- Go to the code directory and run it:
python launch_scientist_bfts.py --load_ideas "ai_scientist/ideas/i_cant_believe_its_not_better.json" --model_writeup "claude-3-5-sonnet-20240620"
- The system generates a JSON file containing the study title and description.
2. Running experiments
- After the idea is generated, the system creates the experiment code (e.g.
experiment.py). - Perform the experiment:
python experiment.py
- The results are saved in the
experimentsfolder in the log, including data and graphs.
3. Writing of papers
- Once the experiment is complete, generate a paper:
python launch_scientist_bfts.py --load_code --add_dataset_ref --model_writeup "o1-preview-2024-09-12" --model_citation "gpt-4o-2024-11-20"
- Outputs LaTeX files, which are stored in the
experiments/timestamp_ideaname/latexFolder. Compile with the LaTeX editor to view it.
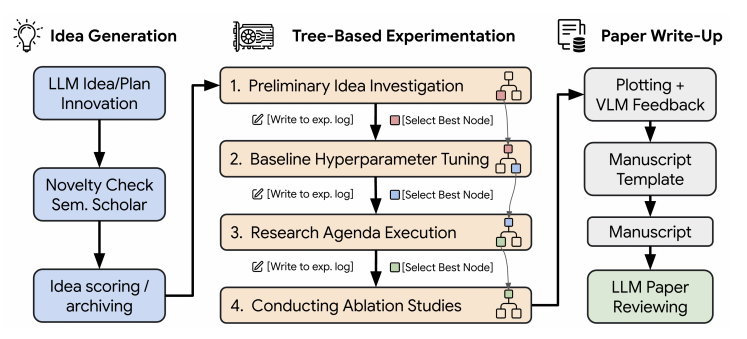
4. Using Agentic Tree Search
- This is a core feature of v2 that optimizes study paths.
- Add parameters at runtime:
python launch_scientist_bfts.py --load_ideas "ai_scientist/ideas/i_cant_believe_its_not_better.json" --tree-search
- generating
unified_tree_viz.htmlIf you have a browser, you can open it to view the search process.
5. Configuration tree search parameters
- compiler
bfts_config.yamlDocumentation: num_workers: Number of nodes for parallel processing, e.g. 3.steps: Maximum number of nodes to explore, e.g. 21.num_drafts: Number of initial research directions.max_debug_depth: Number of debugging attempts.
caveat
- safety: The code executes programs written by the AI and may call dangerous packages or be networked, it is recommended to run it with Docker.
- (manufacturing, production etc) costs: Approximately $15-$20 per experiment, plus $5 for thesis writing.
- success rate: v2 is highly exploratory, has a lower success rate than v1, and is suitable for open research.
- Memory issues: If prompted "CUDA Out of Memory", change small model in JSON file.
These steps give you a complete experience of AI-Scientist-v2's research automation capabilities.
application scenario
- academic research
Researchers use it to validate new algorithms, generate first drafts of papers, and save time. - Educational learning
Students use it to simulate scientific research, generate reports, and learn about experimental design. - technological innovation
Developers use it to test new ideas and quickly generate code prototypes.
QA
- What models are supported?
Support for Claude 3.5 Sonnet, GPT-4o, o1-preview, etc., seellm.pyDocumentation. - How much did the experiment cost?
With Claude 3.5 it's about $15-$20 per session, add $5 for writing. - What should I do if I fail to generate a paper?
The success rate varies depending on the model and complexity of the idea, and the parameters can be adjusted or retried with a different model. - How do I add a new research direction?
existai_scientist/ideas/Add a new JSON file in the directory and modify it with reference to the example.
© Copyright notes
Article copyright AI Sharing Circle All, please do not reproduce without permission.
Related posts

No comments...